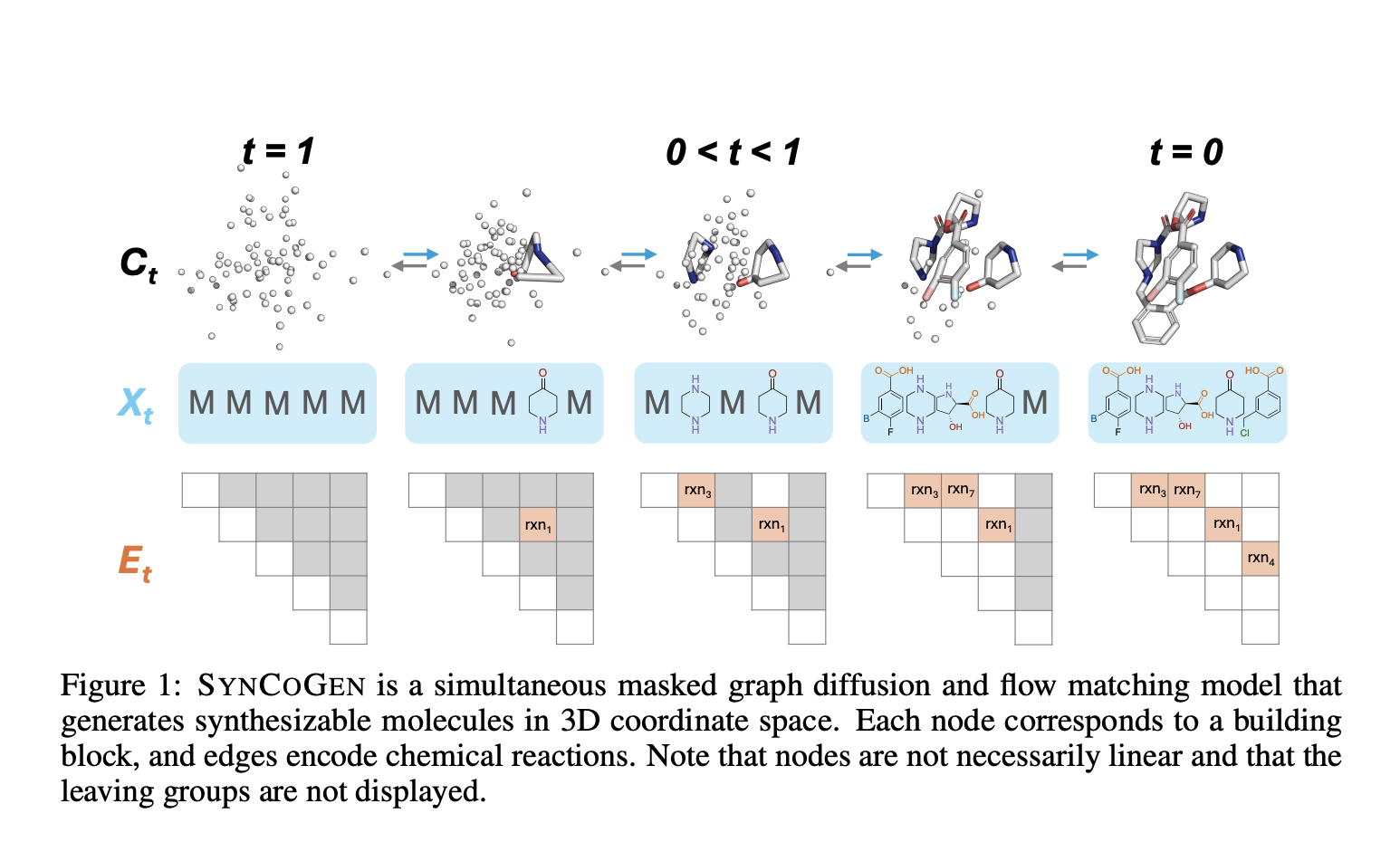
Introduction: the challenge of the generation of synthesible molecules
In the discovery of modern drugs, Generative molecular design models The chemical space available for researchers have considerably widened, allowing rapid exploration of new compounds. However, a major challenge remains: many molecules generated by AI are difficult or impossible to synthesize in the laboratorylimiting their practical value in pharmaceutical and chemical development.
While methods based on models, such as synthetic trees built from reaction models – Adelp send synthetic accessibility, these approaches only capture 2D molecular graphicswithout the rich 3D structural information This determines the behavior of a molecule in biological systems.
3D bridging structure and synthesis: the need for a unified framework
Recent advances in 3D generative models Can generate atomic coordinates directly, allowing a design based on geometry and an improved property prediction. However, most methods do not systematically integrate synthetic feasibility constraints: Resulting molecules can have desired forms or properties, but there is no guarantee that they can be assembled from existing construction blocks using known reactions.
Synthetic accessibility is crucial to succeed Discovery of drugs and the design of materials, causing the need for solutions that simultaneously guarantee both Realistic 3D geometry and direct synthetic roads.


Syncogen: a new frame for the design of synthesable 3D molecules
Researchers from the University of Toronto, the University of Cambridge, McGill University and others have proposed syncogenic (synthesable co-generation) which fills this gap with a pioneer approach which Model jointly both the reaction routes and the atomic coordinates During the generation of molecules. This unified framework allows the generation of 3D molecular structures with Treatable synthetic routesMake sure that each proposed molecule is not only physically significant but also practically synthesable.
Key innovations of syncogen
- Multimodal generation: By mixing Diffusion of the masked graphic (for reaction graphics) with flow correspondence (for atomic coordinates), syncogen samples from the joint distribution of construction blocks, chemical reactions and 3D structures.
- Complete representation of entries: Each molecule is represented as a triple (x, e, c)Or:
- X code the identity of the construction blocks,
- E code for specific reaction types and connection centers,
- C Contains all atomic coordinates.
- Simultaneous training: The graphic and contact details are modeled together, using losses that combine Cross entropy for graphics,, Masked average square error for coordinatesAnd Distance penalties per pair To ensure geometric realism.


The SYSPACE data set: allowing large -scale training and devoted to synthesis
To form the syncogen, the researchers created SystemA set of data with more than 600,000 synthesizable molecules, each built from 93 commercial construction blocks And 19 robust reaction models. Each Syspace molecule is annotated with several Energy minimized 3D conformations (More than 3.3 million structures in total), offering a diversified and reliable training resource which closely reflects the realistic chemical synthesis.
Data construction work flow
- Molecules are systematically built by Iterative reaction assemblyFrom a first building block and choose compatible reaction centers and partners for successive coupling stages.
- For each resulting, multiple molecular graphic low energy complutes are generated and optimized using calculation chemistry methods, ensuring that each structure is both chemically plausible and energetically favorable.
Architecture and training models
Syncogen uses a Semlaflow Backbone, an equivalent neural network (3) originally designed for 3D molecular generation. Architecture includes:
- Specialized exit heads to translate between graphics at the construction blocks And Characteristics at the Atom level.
- Loss functions and non-equilibrium patterns which carefully balance the accuracy of graphics and 3D structural fidelity, including the manipulation of coordinates aware of visibility to take charge of the variable number of atoms and masking.
- Training of innovations such as edges,, compatibility maskingAnd self-order To maintain the generation of chemical valiant molecules.
Performance: of the state of technology leads to a generation of synthesable molecules
Reference
Syncogen reached peak performance On the generation tasks of unconditional 3D molecules, outperforming generative frames of first atom and graphics. Notable improvements include:
- High chemical validity: More than 96% of the molecules generated are chemically valid.
- Higher synthetic accessibility: Retrosynthesis software (Aizynthfinder, Syntheseus) solves rates up to 72%, far exceeding most competing methods.
- Excellent geometric and energetic realism: The confirms generated closely correspond to the length of the connection, the angle and the dihedral distributions of the experimental data sets, with low unrelated interaction energies.
- Practical utility: Syncogen allows a direct generation of synthetic roads In addition to 3D coordinates, only punching computer chemistry and experimental synthesis.
Fragments' link and drug design
Syncogen also demonstrates competitive performance in Integral molecularness for the binding of fragmentsA crucial drug design task. He can generate easily synthesizable analogues complex drugs, producing candidates with favorable mooring scores and retrosynthetic tractability – a feat that is not equaled by conventional generative models.
Future Directions and Applications
Syncogen marks a fundamental advance for conscious molecular generation of synthesisWith potential extensions, in particular:
- Generation conditioned by property: Optimize the desired physicochemical or biological properties directly.
- Protectic pocket conditioning: Generate personalized ligands for liaison sites with specific protein.
- Expansion of reaction space: Incorporate more diverse construction blocks and reaction models to widen the accessible chemical space.
- Automated synthetic robotics: Link of generative models with laboratory automation for the discovery of closed loop drugs and materials.
Conclusion: a step towards the molecular conception of calculation feasible
Syncogen establishes a new reference for Generation of 3D joint molecules and reactionallowing researchers and pharmaceutical scientists to design molecules which are both structurally significant and experimentally feasible. By uniting generative models with strict synthetic constraints, syncogen brings the computer design closer to the realization of laboratory, unlocking new opportunities in Discovery of drugs,, Material scienceand beyond.
FAQ 1: What is syncogen and how does it improve the generation of synthesable 3D molecules?
Syncogen is an advanced generative modeling framework which simultaneously generates 3D structures and synthetic reaction paths for small molecules. By jointly modeling reaction graphics and atomic coordinates, Syncogen guarantees that the generated molecules are not only physically realistic but also easily synthesable in the real laboratory. This double approach allows uniquely the design of practical molecules for the discovery of drugs, filling a critical gap left by previous models which were focused only on 2D structures or the negligence of synthetic accessibility.
FAQ 2: How is syncogen formed to guarantee synthetic accessibility and 3D precision?
Syncogen is formed using the Synspace data set, which includes more than 600,000 synthesable molecules built from a fixed set of reliable construction blocks and reaction models, each associated with multiple 3D complores minimized in energy. The model uses the dissemination of the masked graphic for the reaction graph and the correspondence of the flow for the atomic coordinates, combining the cross entropy of the graph, the average square error coordinates and the distance penalties by pair during training to apply both chemical validity and geometric realism. Training time constraints, such as edge counting limits and compatibility masking, also guarantee the generation of practical and valid molecules.
FAQ 3: What are the main future applications and guidelines for syncogen in chemical and pharmaceutical research?
Syncogen establishes a new standard for the generation of devotable 3D molecules, allowing direct suggestion of synthetic routes alongside 3D structures – key for the design of drugs, the link of fragments and automated synthetic platforms. Future applications include the generation of packaging on specific properties or binding pockets to protein, by widening the library of applicable reactions and building blocks and integration into laboratory robotics for the synthesis and screening of fully automated molecules.
Discover the Paper here. All the merit of this research goes to researchers in this project.
Meet the newsletter of AI dev read by 40K + developers and researchers from Nvidia, Openai, Deepmind, Meta, Microsoft, JP Morgan Chase, Amgen, Aflac, Wells Fargo and 100 others (Subscribe now)
Sajjad Ansari is a last year's first year of the Kharagpur Iit. As a technology enthusiast, he plunges into AI's practical applications by emphasizing the understanding of the impact of AI technologies and their real implications. It aims to articulate complex AI concepts in a clear and accessible way.