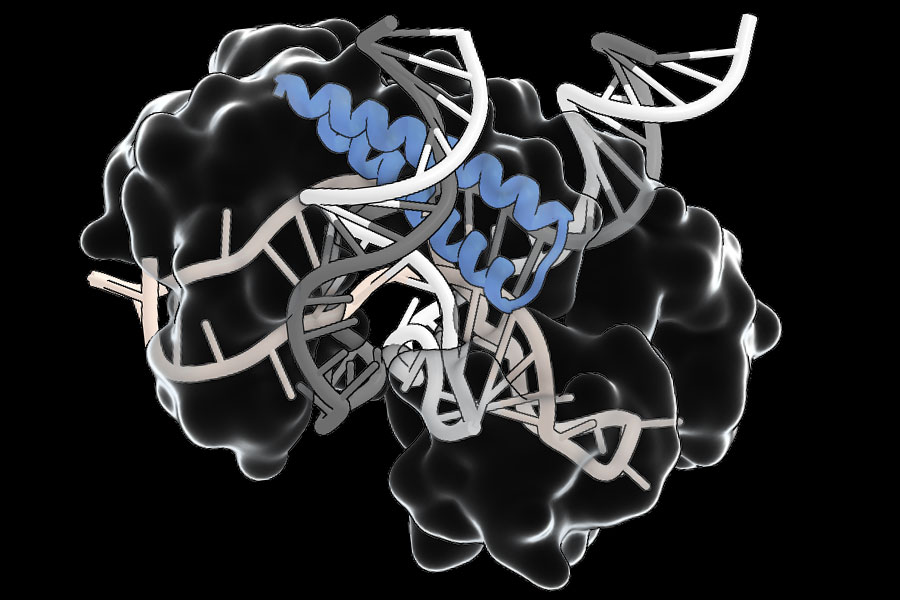
A vast search for natural diversity has led scientists to the McGovern Institute for Brain Research and the Broad Institute of Mit and Harvard to discover old systems with a potential to extend the genome publishing tool box.
These systems, which researchers call the TIGR (RNA Guide RNA prohibited in tandem) systems, use RNA to guide them to specific DNA sites. TIGR systems can be reprogrammed to target any interest sequence of interest, and they have separate functional modules that can act on targeted DNA. In addition to its modularity, the TIGR is very compact compared to other systems guided by RNA, such as CRISPR, which is a major advantage to provide it in a therapeutic context.
These results are reported online on February 27 in the newspaper Science.
“This is a very versatile system guided by RNA with many various features,” explains Feng Zhang, professor of neuroscience at James and Patricia Poitras at MIT, who led the research. The proteins associated with TIGR (CAS) that the Zhang team have found sharing a characteristic RNA link component which interacts with a RNA guide which directs it to a specific genome site. Some have cut DNA on this site, using an adjacent protein adjacent adjacent segment. This modularity could facilitate the development of tools, allowing researchers to exchange new characteristics useful in natural heap proteins.
“Nature is quite incredible,” explains Zhang, who is also an investigator at the McGovern Institute and the Howard Hughes Medical Institute, a basic member of the Broad Institute, a professor of brain and cognitive sciences and biological genius at MIT, and co -director of the K. Lisa Yang and Hock E. Tan Center for molecular therapy at MIT. “It has a huge diversity, and we have explored this natural diversity to find new biological mechanisms and exploit them for different applications to manipulate biological processes,” he says. Previously, Zhang's team has adapted Bacterial Crispr systems into genes of genes of genes that have transformed modern biology. His team also found a variety of programmable proteins, both CRISPR systems and beyond.
In their new work, to find new programmable systems, the team started with zero in a structural characteristic of the CRISPR-CAS9 protein which links to the enzyme RNA guide. This is a key characteristic that made Cas9 such a powerful tool: “Being guided by RNA makes reprogramming relatively easy, because we know how RNA is linked to other DNAs or other RNAs”, explains Zhang. His team searched hundreds of millions of organic proteins with known or predicted structures, looking for who shared a similar area. To find more distant proteins, they used an iterative process: from Cas9, they identified a protein called IS110, which had previously shown that others are linked to RNA. They then focused on the structural characteristics of the IS110 which allow the link to the RNA and repeated their research.
At this stage, research had revealed so much distant protein that they turned to artificial intelligence to give meaning to the list. “When you make an iterative and deep mine, the resulting tubes can be so diverse that they are difficult to analyze using standard phylogenetic methods, which depend on the preserved sequence,” explains Guilhem Faure, computer biologist in the Zhang laboratory. With a large protein language model, the team was able to group the proteins they had found in groups according to their probable evolutionary relationships. A group separated from the others, and its members were particularly intriguing because they were coded by genes with regularly spaced repetitive sequences recalling an essential component of CRISPR systems. These are tigr-tas systems.
Zhang's team has discovered more than 20,000 different heap proteins, especially in viruses infected with bacteria. The sequences in the repetitive region of each gene – its Tigr paintings – code for a RNA guide which interacts with the binding part of the protein RNA. In some, the RNA liaison region is adjacent to a section of the DNA of the protein. Others seem to be linked to other proteins, which suggests that they could help direct these proteins to DNA targets.
Zhang and his team have experienced dozens of heap proteins, demonstrating that some can be programmed to make cut for DNA in human cells. While they plan to develop tigr-tas systems in programmable tools, researchers are encouraged by features that could make these tools particularly flexible and precise.
They note that CRISPR systems can only be directed to DNA segments which are flanked by short patterns called PAM (Protospace the adjacent patterns). Tig Tas proteins, on the other hand, do not have such a requirement. “This theoretically means that any genome site should be targeting,” said scientific advisor Rhiannon Macrae. The team's experiences also show that TIGR systems have what Faure calls a “double guide system”, interacting with the two strands of the DNA double propeller at home on their target sequences, which should ensure that they only act where they are led by their RNA guide. In addition, heap proteins are compact – a quarter of the size CAS9, on average – makes them easier to deliver, which could overcome a major obstacle to the therapeutic deployment of gene editing tools.
Excited by their discovery, Zhang's team is now studying the natural role of tig systems in viruses, as well as the way they can be adapted to research or therapy. They determined the molecular structure of one of the Pile proteins they have found in human cells, and will use this information to guide their efforts to make it more effective. In addition, they note connections between tigr-tas systems and certain RNA processing proteins in human cells. “I think there is more there to study in terms of what some of these relationships can be, and this can help us better understand how these systems are used in humans,” explains Zhang.
This work was supported by Helen Hay Whitney Foundation, Howard Hughes Medical Institute, K. Lisa Yang and Hock E. Tan Center for Molecular Therapeutics, Broad Institute Programmable Therapeutics Gift Donors, Pershing Square Foundation, William Ackman, Neri Oxman, The Phillips Family, J. The Charitable BT Foundation.